- 快召唤伙伴们来围观吧
- 微博 QQ QQ空间 贴吧
- 文档嵌入链接
- 复制
- 微信扫一扫分享
- 已成功复制到剪贴板
Automatic Image Annotation - Department of Knowledge Technologies
展开查看详情
1 .Ivica Dimitrovski 1 , Dragi Kocev 2 , Suzana Lo skovska 1 , Sašo Džeroski 2 1 Faculty of Electrical Engineering and Information Technologies, Department of Computer Science, Skopje, Macedonia 2 Jožef Stefan Institute, Department of Knowledge Technologies, Ljubljana, Slovenia Hierarchical Classification of Diatom Images using Predictive Clustering Trees
2 .Outline Hierarchical multi-label classification system for diatom image classification Contour and feature extraction from images Predictive Clustering Trees Ensembles : Bagging and random forests Experimental Design Results and Discussion
3 .Diatom image classification (1) Diatoms: large and ecologically important group of unicellular or colonial organisms (algae) Variety of uses: water quality monitoring, paleoecology and forensics
4 .Diatom image classification (2) 200000 different diatom species, half of them still undiscovered Automatic diatom classification image processing (feature extraction from images) image classification (labels and groups the images) Labels can be organized in a hierarchy and an image can be labeled with more than one label Predict all different levels in the hierarchy of taxonomic ranks: genus, species, variety, and form Goal of the complete system: assist a taxonomist in identifying a wide range of different diatoms
5 .Diatom image classification (3) Set of images with their visual descriptors and annotations Taxonomic rank with hierarchical structure
6 .Contour extraction from images Pre-segmentation of an image separate the diatom objects from dark or light debris identify the regions with structured objects merge nested regions Edge-based thresholding for contour extraction locate the boundary between the objects and the background produce a binary (black and white) image with the diatom contours Contour following trace the region borders in the binary image
7 .Feature extraction from images Simple geometric properties length, width, size and the length-width ratio Simple shape descriptors rectangularity, triangularity, compactness, ellipticity, and circularity Fourier descriptors 30 coefficients SIFT histograms Invariant to changes in illumination, image noise, rotation, scaling, and small changes in viewpoint
8 .Predictive Clustering Trees (PCTs) Standard Top-Down Induction of DTs Hierarchy of clusters Distance measure: minimization of intra-cluster variance Instantiation of the variance for different tasks Multiple targets, sequences, hierarchies
9 .CLUS System where the PCTs framework is implemented (KULeuven & JSI) Available for download at http://www.cs.kuleuven.be/~dtai/clus The top-down induction algorithm for PCTs: Selecting the tests: reduction in variance caused by partitioning the instances 9
10 .PCTs for Hierarchical Multi-Label Classification HMLC: an example can be labeled with multiple labels that are organized in a hierarchy { 1, 2, 2.2 }
11 .PCTs for Hierarchical Multi-Label Classification Variance: average squared distance between each example’s label and the set’s mean label Weighted Euclidean distance: an error at the upper levels costs more than an error at the lower levels
12 .Ensemble methods Ensembles are a set of predictive models Unstable base classifiers Voting schemes to combine the predictions into a single prediction Ensemble learning approaches Modification of the data Modification of the algorithm Bagging Random Forest
13 .Ensemble methods Image from van Assche, PhD Thesis, 2008 Training set … 1 2 n 3 (Randomized) Decision tree algorithm n classifiers … Test set L 1 L 2 L 3 L n n predictions vote n bootstrap replicates L (Randomized) Decision tree algorithm (Randomized) Decision tree algorithm (Randomized) Decision tree algorithm Training set … 1 2 n 3 (Randomized) Decision tree algorithm n classifiers … Test set L 1 L 2 L 3 L n n predictions vote n bootstrap replicates L (Randomized) Decision tree algorithm (Randomized) Decision tree algorithm (Randomized) Decision tree algorithm Training set … 1 2 n 3 (Randomized) Decision tree algorithm n classifiers … Test set L 1 L 2 L 3 L n n predictions vote n bootstrap replicates L (Randomized) Decision tree algorithm (Randomized) Decision tree algorithm (Randomized) Decision tree algorithm Training set … 1 2 n 3 (Randomized) Decision tree algorithm n classifiers … Test set L 1 L 2 L 3 L n n predictions vote n bootstrap replicates L (Randomized) Decision tree algorithm (Randomized) Decision tree algorithm (Randomized) Decision tree algorithm
14 .Ensemble methods Image from van Assche, PhD Thesis, 2008 Training set … 1 2 n 3 (Randomized) Decision tree algorithm n classifiers … Test set L 1 L 2 L 3 L n n predictions vote n bootstrap replicates L (Randomized) Decision tree algorithm (Randomized) Decision tree algorithm (Randomized) Decision tree algorithm Training set … 1 2 n 3 (Randomized) Decision tree algorithm n classifiers … Test set L 1 L 2 L 3 L n n predictions vote n bootstrap replicates L (Randomized) Decision tree algorithm (Randomized) Decision tree algorithm (Randomized) Decision tree algorithm Training set … 1 2 n 3 (Randomized) Decision tree algorithm n classifiers … Test set L 1 L 2 L 3 L n n predictions vote n bootstrap replicates L (Randomized) Decision tree algorithm (Randomized) Decision tree algorithm (Randomized) Decision tree algorithm Training set … 1 2 n 3 CLUS n classifiers … Test set L 1 L 2 L 3 L n n predictions vote n bootstrap replicates L CLUS CLUS CLUS
15 .ADIAC diatom image database Three different subset of images: 1099 images classified in 55 different taxa 1020 images classified in 48 different taxa 819 images classified in 37 different taxa The diatoms vary in shape and ornamentation
16 .Experimental design – classifier Random Forests and Bagging of PCTs for HMLC: Feature Subset Size: 10% of the number of descriptive attributes Number of classifiers: 100 un-pruned trees Combine the predictions output by the base classifiers: probability distribution vote
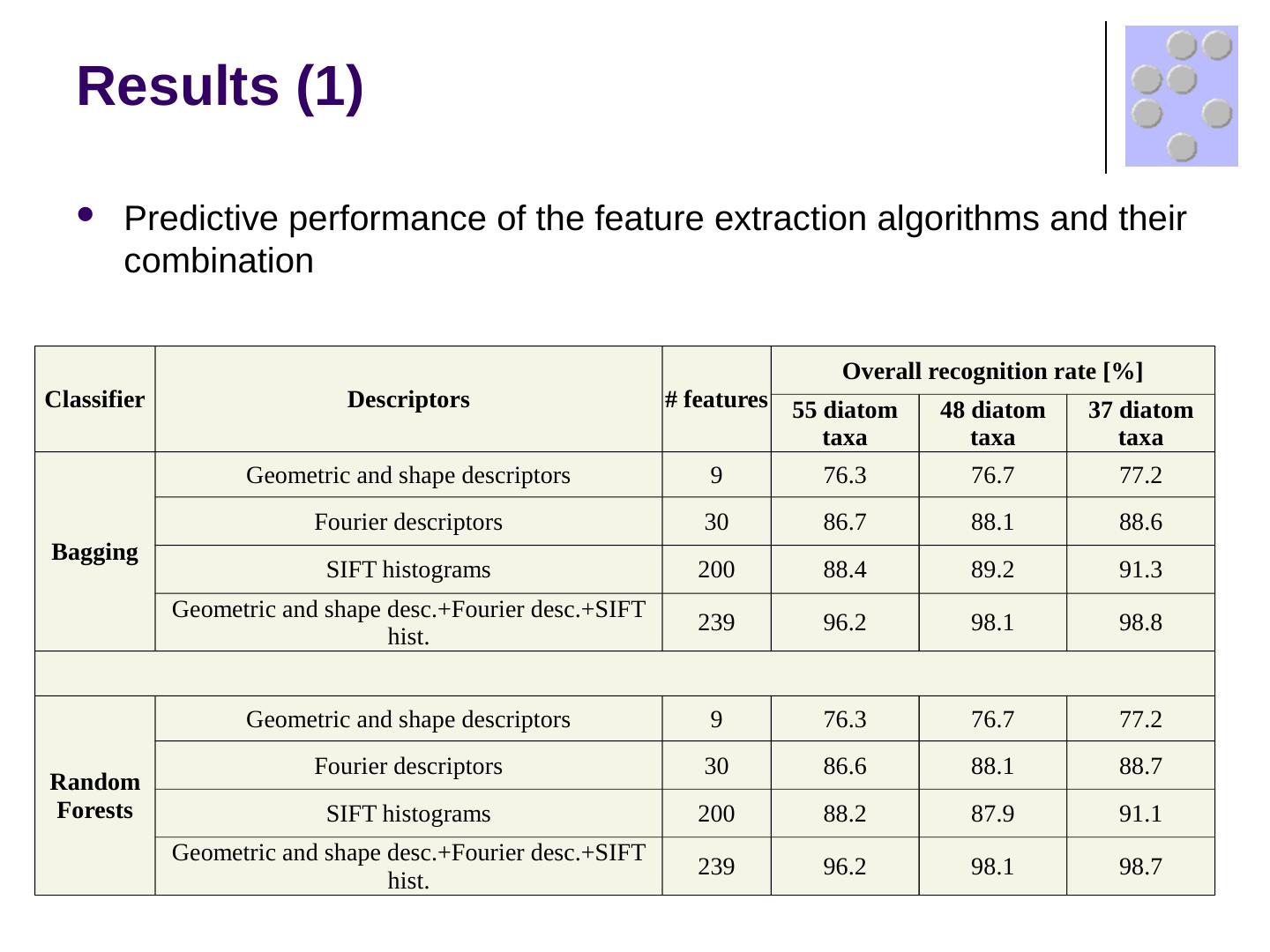
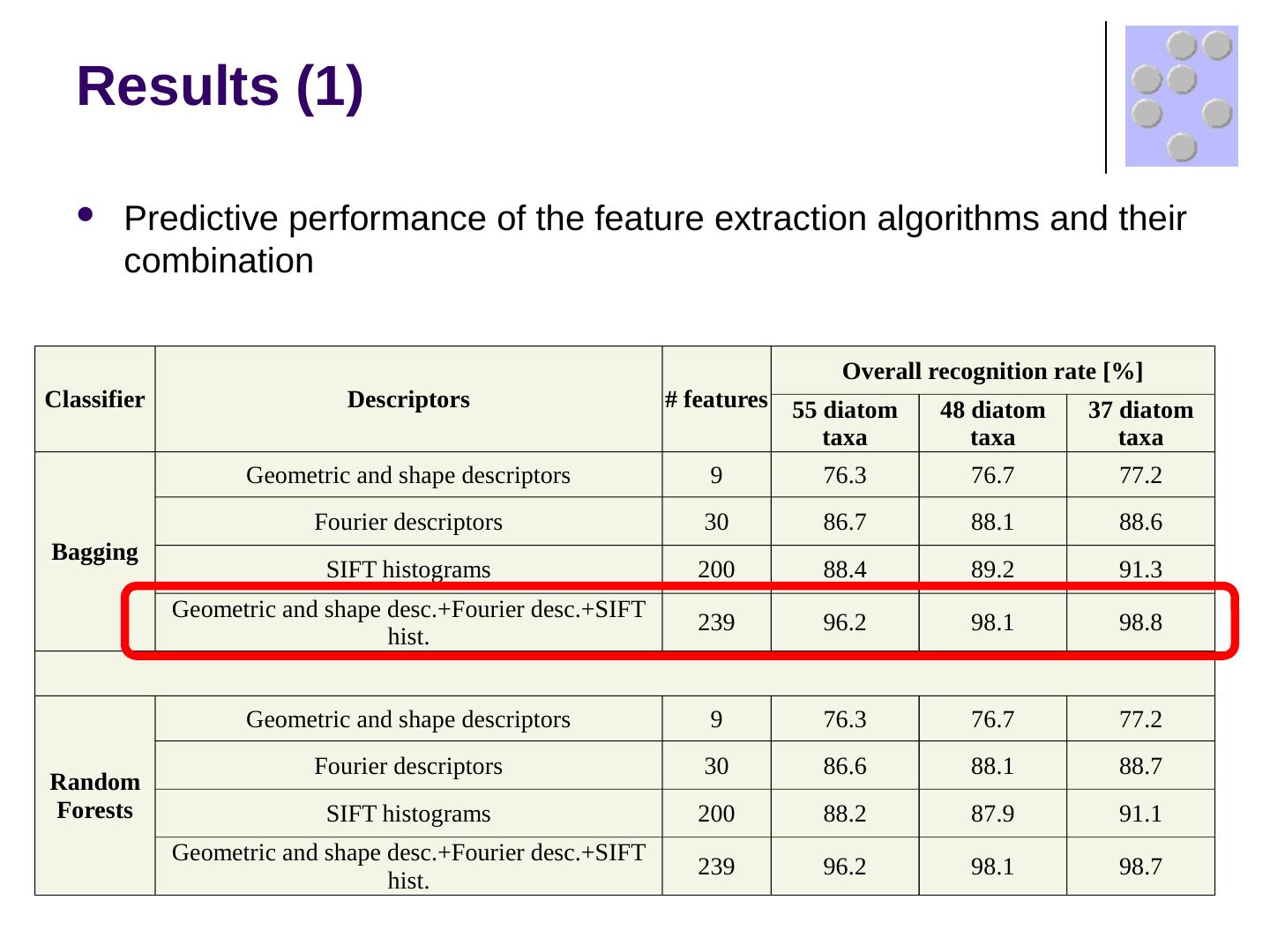
17 .Results (1) Predictive performance of the feature extraction algorithms and their combination Classifier Descriptors # features Overall recognition rate [%] 55 diatom taxa 48 diatom taxa 37 diatom taxa Bagging Geometric and shape descriptors 9 76.3 76.7 77.2 Fourier descriptors 30 86.7 88.1 88.6 SIFT histograms 200 88.4 89.2 91.3 Geometric and shape desc.+Fourier desc.+SIFT hist. 239 96.2 98.1 98.8 Random Forests Geometric and shape descriptors 9 76.3 76.7 77.2 Fourier descriptors 30 86.6 88.1 88.7 SIFT histograms 200 88.2 87.9 91.1 Geometric and shape desc.+Fourier desc.+SIFT hist. 239 96.2 98.1 98.7
18 .Results (1) Predictive performance of the feature extraction algorithms and their combination Classifier Descriptors # features Overall recognition rate [%] 55 diatom taxa 48 diatom taxa 37 diatom taxa Bagging Geometric and shape descriptors 9 76.3 76.7 77.2 Fourier descriptors 30 86.7 88.1 88.6 SIFT histograms 200 88.4 89.2 91.3 Geometric and shape desc.+Fourier desc.+SIFT hist. 239 96.2 98.1 98.8 Random Forests Geometric and shape descriptors 9 76.3 76.7 77.2 Fourier descriptors 30 86.6 88.1 88.7 SIFT histograms 200 88.2 87.9 91.1 Geometric and shape desc.+Fourier desc.+SIFT hist. 239 96.2 98.1 98.7
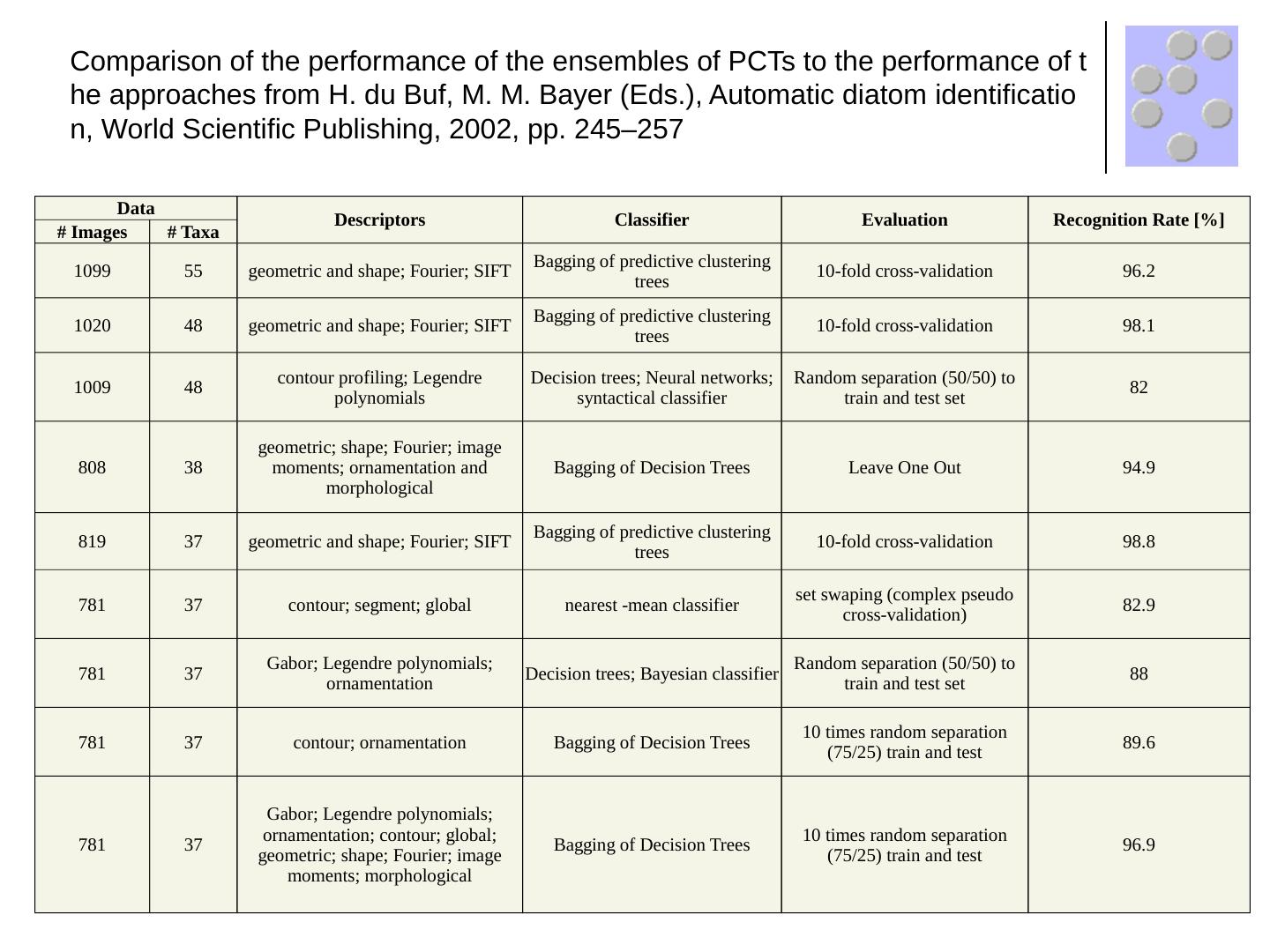
19 .Data Descriptors Classifier Evaluation Recognition Rate [%] # Images # Taxa 1099 55 geometric and shape; Fourier; SIFT Bagging of predictive clustering trees 10-fold cross-validation 96.2 1020 48 geometric and shape; Fourier; SIFT Bagging of predictive clustering trees 10-fold cross-validation 98.1 1009 48 contour profiling; Legendre polynomials Decision trees; Neural networks; syntactical classifier Random separation (50/50) to train and test set 82 808 38 geometric; shape; Fourier; image moments; ornamentation and morphological Bagging of Decision Trees Leave One Out 94.9 819 37 geometric and shape; Fourier; SIFT Bagging of predictive clustering trees 10-fold cross-validation 98.8 781 37 contour; segment; global nearest -mean classifier set swaping (complex pseudo cross-validation) 82.9 781 37 Gabor; Legendre polynomials; ornamentation Decision trees; Bayesian classifier Random separation (50/50) to train and test set 88 781 37 contour; ornamentation Bagging of Decision Trees 10 times random separation (75/25) train and test 89.6 781 37 Gabor; Legendre polynomials; ornamentation; contour; global; geometric; shape; Fourier; image moments; morphological Bagging of Decision Trees 10 times random separation (75/25) train and test 96.9 Comparison of the performance of the ensembles of PCTs to the performance of the approaches from H. du Buf , M. M. Bayer (Eds.), Automatic diatom identification, World Scientific Publishing, 2002, pp. 245–257
20 .Data Descriptors Classifier Evaluation Recognition Rate [%] # Images # Taxa 1099 55 geometric and shape; Fourier; SIFT Bagging of predictive clustering trees 10-fold cross-validation 96.2 1020 48 geometric and shape; Fourier; SIFT Bagging of predictive clustering trees 10-fold cross-validation 98.1 1009 48 contour profiling; Legendre polynomials Decision trees; Neural networks; syntactical classifier Random separation (50/50) to train and test set 82 808 38 geometric; shape; Fourier; image moments; ornamentation and morphological Bagging of Decision Trees Leave One Out 94.9 819 37 geometric and shape; Fourier; SIFT Bagging of predictive clustering trees 10-fold cross-validation 98.8 781 37 contour; segment; global nearest -mean classifier set swaping (complex pseudo cross-validation) 82.9 781 37 Gabor; Legendre polynomials; ornamentation Decision trees; Bayesian classifier Random separation (50/50) to train and test set 88 781 37 contour; ornamentation Bagging of Decision Trees 10 times random separation (75/25) train and test 89.6 781 37 Gabor; Legendre polynomials; ornamentation; contour; global; geometric; shape; Fourier; image moments; morphological Bagging of Decision Trees 10 times random separation (75/25) train and test 96.9 Comparison of the performance of the ensembles of PCTs to the performance of the approaches from H. du Buf, M. M. Bayer (Eds.), Automatic diatom identification, World Scientific Publishing, 2002, pp. 245–257
21 .Results (2) The presented approach has very high predictive performance (ranging from 96.2% to 98.7%) Recognition rates of 100% for the majority of taxa Lower recognition rates are achieved for taxa that are very similar to each other and difficult to distinguish Eunotia diatoms (presented on image) , Fallacia diatoms Our results are better than the ones obtained from human annotators (63.3% recognition rate)
22 .Conclusion Novel approach to taxonomic identification of taxa from microscopic images Different feature extraction approaches and hierarchical multi-label classification Very high predictive performance - the best reported performance on this dataset