- 快召唤伙伴们来围观吧
- 微博 QQ QQ空间 贴吧
- 文档嵌入链接
- 复制
- 微信扫一扫分享
- 已成功复制到剪贴板
轻量级深度学习物联网设备用于低功耗便携式皮肤癌检测
展开查看详情
1 . Apply Lightweight Deep Learning on Internet of Things for Low-Cost and Easy-To-Access Skin Cancer Detection Pranjal Sahua , Dantong Yub , and Hong Qina a StonyBrook University, NY, USA b New Jersey Institute Of Technology, NY, USA ABSTRACT Melanoma is the most dangerous form of skin cancer that often resembles moles. Dermatologists often rec- ommend regular skin examination to identify and eliminate Melanoma in its early stages. To facilitate this process, we propose a hand-held computer (smart-phone, Raspberry Pi) based assistant that classifies with the dermatologist-level accuracy skin lesion images into malignant and benign and works in a standalone mobile device without requiring network connectivity. In this paper, we propose and implement a hybrid approach based on advanced deep learning model and domain-specific knowledge and features that dermatologists use for the inspection purpose to improve the accuracy of classification between benign and malignant skin lesions. Here, domain-specific features include the texture of the lesion boundary, the symmetry of the mole, and the boundary characteristics of the region of interest. We also obtain standard deep features from a pre-trained network optimized for mobile devices called Google’s MobileNet. The experiments conducted on ISIC 2017 skin cancer classification challenge demonstrate the effectiveness and complementary nature of these hybrid features over the standard deep features. We performed experiments with the training, testing and validation data splits provided in the competition. Our method achieved area of 0.805 under the receiver operating characteristic curve. Our ultimate goal is to extend the trained model in a commercial hand-held mobile and sensor device such as Raspberry Pi and democratize the access to preventive health care. Keywords: Skin cancer, Melanoma, deep learning, transfer learning, Internet of things, Raspberry Pi, Polar Harmonic Transform 1. INTRODUCTION Skin cancer is a common form of cancer in the United States. Melanoma is the deadliest form of skin cancer and not only has the number of the skin cancer cases increased between the period 2002-2006 and 2007-2011, but the cost of treatment has increased substantially as well.1 According to an estimate in 2017, there will be 87,110 new cases of melanoma in the United States and 9,730 deaths from the disease. Melanoma cancer in an advanced stage might invade internal organs, such as lungs via blood vessels, making the treatment even harder. However, Melanoma is curable if identified in early stages and a regular skin examination prescribed by Dermatologists serves the purpose of early diagnosis. Dermatologists employ digital imaging for identifying and diagnosing skin cancer. Melanoma skin cancer has certain characteristic appearances: Asymmetric mole on skin (A), irregularity in the border (B), multi and dark color (C), diameter of the mole (D), and evolving size (E), which in short are denoted as the “ABCDE” rule for identifying melanoma. Figure 1 shows some images that explains the “ABCDE” rule. There are different computer-aided approaches in the past to extract features and automate the identification of melanoma from skin images, for example, features encoding the ABCD rule such as circularity,2 the Histogram Of Oriented Gradients features (hog) used with an SVM classifier,3 the 3D shape features obtained from the reconstructed 3D shape of lesion from images,4 the texture features5 of contrast, correlation, and homogeneity6 for a neural network classifier. Additional work includes a bag of features using the histogram of codewords7 and an ensemble of global and local texture features.8 Mostly all of these work perform experiments on small and Further author information: Pranjal Sahu: E-mail: psahu@cs.stonybrook.edu
2 . Symmetric Smooth border One color Smaller diameter Smaller size initially Asymmetric Irregular border Multi color Larger diameter Growth in size Figure 1: The images demonstrating the ABCDE rule for identifying melanoma. The first row comprises benign lesions and the second row shows the Melanoma samples.9 private datasets. Therefore it is not simple to compare the effectiveness of these approaches. Recent success in deep learning has shown that a data-driven approach outperforms the one based on the hand-crafted features for computer vision tasks such as image classification and segmentation. However, training deep networks requires a significant amount of training data, which is typically not available for the biomedical domain. To raise awareness and to obtain the state-of-the-art methods required for identifying melanoma skin cancer, International Skin Cancer Imaging Collaboration (ISIC) organized the ISBI 2017 challenge “Skin Lesion Analysis Towards Melanoma Detection”. The challenge provided a dataset comprising around 2700 images along with the annotation by domain experts. A small amount of training data is not suitable for training a deep network from scratch. Moreover, training such a deep model requires high-performance computing resources (for instance GPU). To meet the computing/data-intensive requirements, many applications often adopted a cloud-based approach in which deep neural models are trained and hosted on remote clouds as infrastructure-as-a-service (Iaas). For actual interference, this method relies on high-speed networks to transfer input data and inference results back from/to users. However, limited internet access and data privacy prohibit the cloud-based solution from gaining mass popularity. A vast majority of the population still does not have access to the Internet. For instance, only 26% of Indian population has access to the Internet.9 This challenge calls for a method that is not only cost-effective but also independent of the Internet. Furthermore, data privacy is another challenge that requires no personal biometric data be exposed to the third party without patients’ permission. Therefore a method needs to avoid data transfer which renders the cloud-based solution inappropriate. To mitigate these problems and bring the benefit of artificial intelligence aided diagnosis system to the rural and remote region where the Internet connectivity is limited and even not available, we propose an in-mobile portable device to identify melanoma skin cancer that utilizes hybrid features extracted from images. The hybrid features comprise the handcrafted shape representation of “A(symmetrical)” and “B(order)” characteristics of lesion and the feature maps generated by a pre-trained deep network model, called MobileNet.10 We apply the transfer learning mechanism where MobileNet is first trained on GPU platform and is then fine-tuned on the mobile platform, such as Raspberry Pi. We train the prediction model with hybrid features on a low-cost Raspberry Pi. This process altogether has the advantage of incorporating new data into the deep learning prediction model without relying on high-performance supercomputers. Furthermore, edge computing has been facilitated by the introduction of low-power deep learning inference accelerators such as Movidius Neural Compute Stick11 using which a deep neural network model can be deployed on small devices such as Raspberry Pi. This expedites the data collection process as well and leads to a better prediction model later. Our contributions in this paper include: • Presenting a hybrid approach utilizing deep feature maps extracted from the MobileNet network and shape features encoded by a 2-D Polar Harmonic Transform descriptor for Melanoma classification.
3 . • Training the prediction model and performing the inference on a Raspberry PI (IoT device). • Evaluating the performance of Training and Inference on Raspberry PI. Figure 2 shows the framework of incremental light-weight deep learning and real-time inference. In the first step, we will pre-train the MobileNet CNN with a large amount data in ImageNet12 on HPC and GPU. Secondly, the trained model is migrated from high-performance GPU to the edge computing and Internet of Things (IoT), including Raspberry PI and smartphones. We will train the prediction model using the top-level fully-connected features obtained by feeding skin image data. Because the IoT device is reserved for image preprocessing and the other task of incremental learning and training when new skin images are available, we have to off-load the feature extraction task to dedicated hardware for fast and accurate real-time prediction. In Figure 2, we adopted the energy-efficient Intel MovidiusT M MyriadT M VPU processors for the offloaded task. Our preliminary study13 confirmed that the speed of using GoogleLeNet to classify 224x224 skin images on Movidius Neural Compute Stick (NCS) (0.65s/image) is five times as fast as that of Raspberry Pi (3.3s/image) and meets the requirement of sub-second prediction. Therefore, we will deploy the MobileNet model into the Intel Movidius NCS, a system on chip (SoC), for accelerated inferences and predictions. In Section 2 we first describe our training procedure on Raspberry Pi using hybrid features. We demonstrate that hybrid features perform better than only black box deep feature maps in Section 3, and finally conclude and discuss the future directions for improvement. Figure 2: Our proposed framework shows the training of Deep network on clouds. Transfer learning migrates the pre-trained network to an IoT device and actual inference requires no need of network connectivity. 2. METHOD DESCRIPTION Because the ISIC dataset does not provide the history and diameter of a lesion, we can only use the ABC (Asymmetry, Border irregularity, Color) characteristics of a lesion. The color characteristics are obtained using deep features. To obtain shape features, we first need to preprocess input images and then apply the Polar Harmonic Transform (PHT). In the preprocessing stage, an image is first converted to the YCbCr color space. Then the image is resized to 56x56 to reduce the computational complexity. We only use the luminance (Y) channel to obtain shape features. Histogram equalization is performed on the (Y) luminance channel to enhance contrast. Finally, we perform the Polar Harmonic Transform on the processed image to obtain the features that encode the border irregularity and asymmetry of a lesion. Then we feed the combined feature vector to an SVM (Support Vector Machine) classifier for training and inference. Figure 3 shows the pipeline of feature extraction and modeling training. In the following sections, we will describe our method in details. 2.1 Deep features extraction Deep learning has revolutionized the domains of computer vision and image processing. The Inception Net trained on ImageNet and fined tuned with transfer learning on skin images achieved the dermatologist level accuracy in classifying skin lesion and detecting skin cancer.14 Recently Google introduced a set of deep networks based on
4 .Figure 3: The pipeline of the model training and inference using hybrid features. We extract deep features using MobileNet and concatenate them with the shape descriptors extracted using the Polar Harmonic Transform. We then train an SVM on the obtained hybrid feature vector. depthwise separable filters for mobile devices, called MobileNets that paved the road to migrate complex network models to mobile and portable devices. In this paper, we choose Raspberry PI 3 model B because it has low cost, supports camera and display devices and comes with a quad-core processor and 1 GB RAM. These tight resource constraints on RAM and processing power necessitates a light-weight deep network model and therefore, we chose MobileNets in our experiments instead of full-fledged deep learning networks, such as Inception V315 or ResNet.16 Table 1 compares the number of parameters and size of the Inception V3 model with a MobileNet model. We first reshape the resolution of all images to 224x224, each of which has three channels and then feed them into the MobileNet to obtain deep features from the penultimate layer that have the dimensionality of 1001. Table 1: Comparing the size and number of parameters between two deep network models Deep Network Model Number of parameters Size of model Memory Usage (in MB) MobileNet v1 1.0 224 4.24 million 72 MB 17 Inception V3 24 million 92 MB 100 2.2 Polar Harmonic Transform Descriptors Our main motive is to run the classification/prediction model on devices with moderate computational power, for example, Raspberry Pi, therefore the model and its associated method must be lightweight in term of computation and yet robust in term of accuracy. We choose to use the Polar Harmonic Transform (PHT)17 to obtain shape features since it satisfies all these requirements. In our experiments, we use PHT to extract the the radial and angular descriptors that encode the border irregularity and asymmetry of a skin lesion. It has been demonstrated that the PHT can reconstruct the original image given a sufficient number of coefficients17 therefore its derived features are discriminative and powerful enough to encode the shape characteristics of the image. Figure 5 shows example of reconstructed grayscale image using PHT. The PHT takes its original form of Fourier Transform and applies the transform in polar coordinates. Given a grayscale luminance channel image obtained after the preprocessing stage, we use f (x, y) to denote the inten-
5 .sity at the pixel location (x, y). We reduce the continuous PHT into the discrete Fourier series (basis), apply the discrete PHT transform with respect to the basis functions ψp,q (x, y) (Equation 1) and attain an image’s frequency representation of the order (p, q) ∈ Z 2 . The transformed coefficients Mp,q are defined as follows: ∗ Mp,q = ψp,q (x, y)f (x, y)∆x∆y, (1) y x where ∗ denotes the complex conjugate. Because our goal is to discover the characteristics of border (ir)regularity and (a)symmetry, the Fourier Transform in the polar coordinates is more effective than the one in the Cartesian coordinates in detecting rotation (a)symmetry.18 The conversion between two coordinate systems is straightfor- ward: ψp,q (x, y) = ψp,q (r, φ), where r = x2 + y 2 and φ = arctan(y/x) with the center of the circle at (0, 0). To ease the complexity of analysis and extract simple features, we need to decompose the original images into the basic wave patterns with clear and separate radial and angular components. To ensure these, the selected harmonic basis functions must possess two properties: the first one is the base functions must be mutually orthogonal to each other,17, 19 and the second one is they should take the form of separation of variables, i.e. ψp,q (r, φ) = Rp (r)Φq (φ), where Rp (r) is the radial component in which only variable r occurs and Φq (φ) is the angular moment defined on the single variable φ. The radial and angular moments are defined as follows: 2 Rp (r) = ei2πpr , (2) iqψ Φq (ψ) = e , (3) where (p, q) ∈ Z 2 . We also used two other radial kernels that are defined as follows: RpC (r) = cos(πpr2 ) , (4) RpS (r) = sin(πpr2 ) . (5) We used three transforms in total to obtain the shape descriptors. The three descriptors are differentiated by their radial components, i.e. the Polar Complex Exponential Transform (PCET) Rp (r), Polar Cosine Transform (PCT) RpC (r) and Polar Sine Transform (PST) RpS (r). The kernel computation of PHT is straightforward and does not suffer from numerical instability. The precomputed kernels can be stored for late use during the inference step and need not be recalculated every time. Moreover, the magnitude of coefficients, |Mp,q |, are rotation-invariant.19 We experimented with |p| <= C and |q| <= C where, C ∈ [5, 10, 15, 20]. Finally, we concatenate all |Mp,q | coefficients to obtain shape descriptor. For instance, when C = 15 the total length of the shape descriptor is 3 × 31 × 31 = 2883. 2.3 Support Vector Machine for classification For the final classification task, we used support vector machines (SVM). Mobilenet and shape features obtained using PHT are concatenated to create hybrid features. We performed a three-way data split, i.e., training- validation-testing and used the validation set to select the kernel for SVM. We experimented with four types of kernels, namely, linear, polynomial, sigmoid and radial basis function. The ISIC 2017 competition dataset comprised of 2000, 619 and 131 images in the splits of training, testing and validation respectively. The dataset is unbalanced because it contains 494 melanoma and 2256 benign samples in total. Therefore we train the SVM with the class weights such that weights for each class are inversely proportional to the class frequency in the training data. In Figure 6, we showed the ROC curves with the four kernels on the validation set and using the hybrid features. We observed that the radial basis function kernel performed the best, and thereby proceeded with this kernel. 3. RESULTS We first demonstrate the improvement in performance by using the hybrid features in Section 3.1. In Section 3.2 we compare the time complexity requirement for training and inference of two models on Raspberry Pi (1) Using only the bottleneck features from the deep neural network and (2) Using hybrid ones.
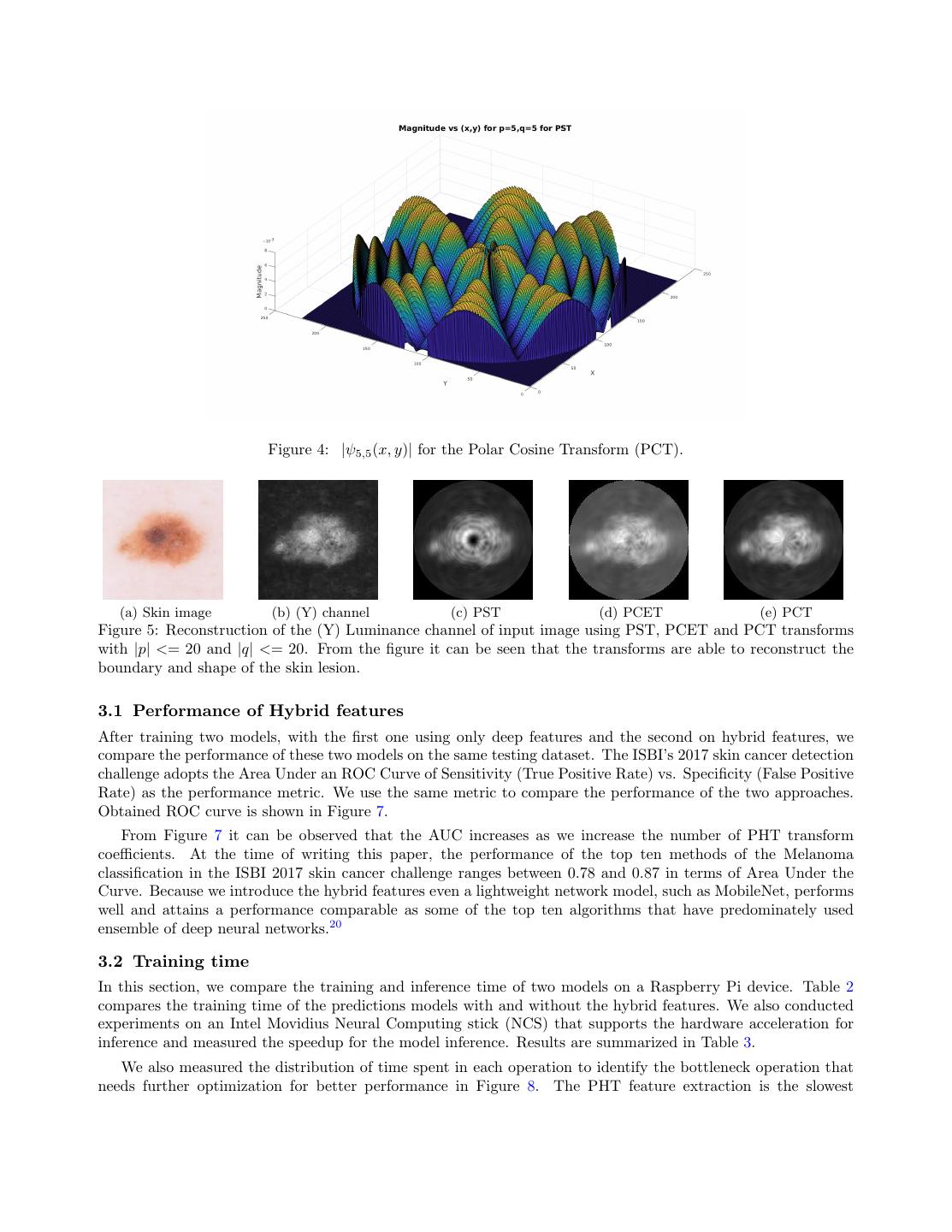
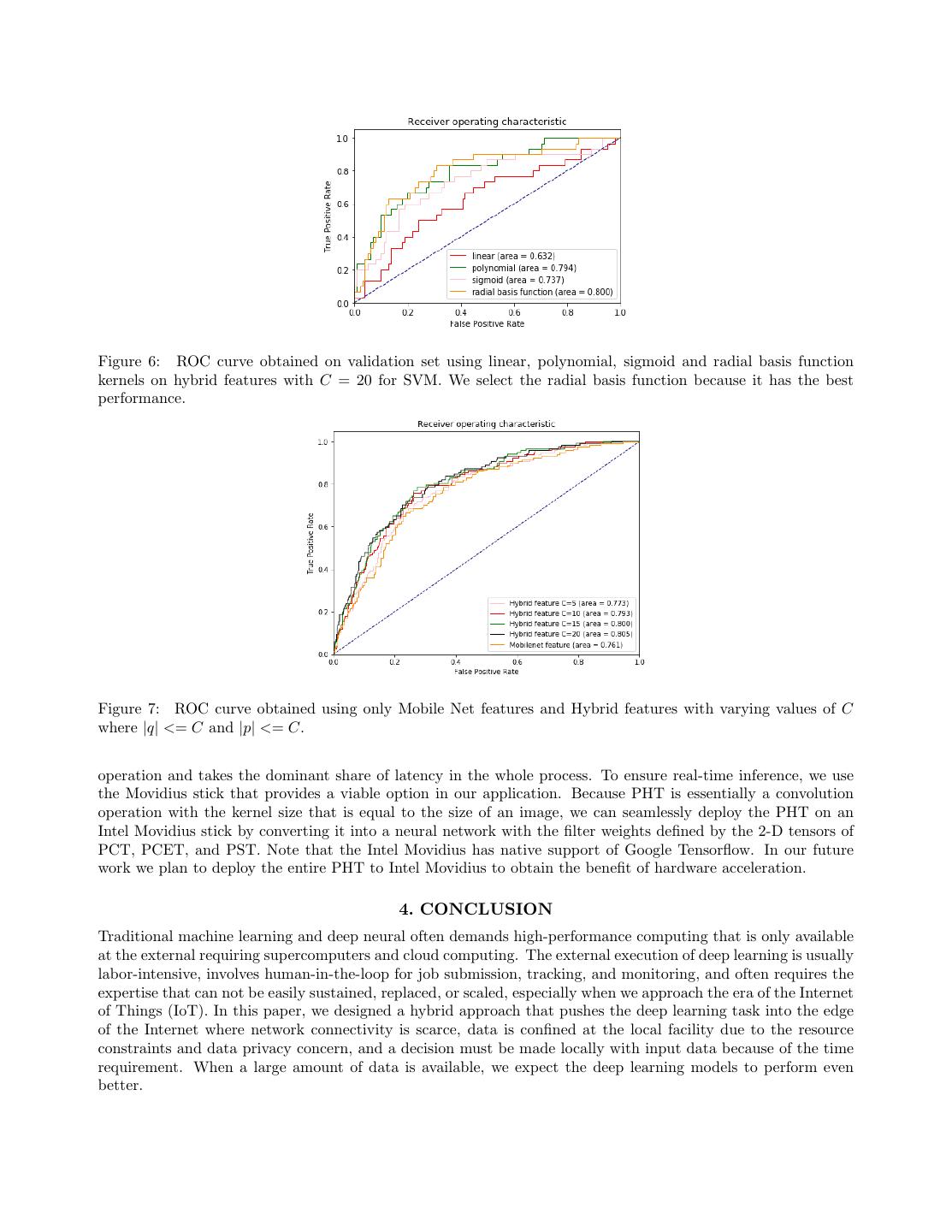
6 . Figure 4: |ψ5,5 (x, y)| for the Polar Cosine Transform (PCT). (a) Skin image (b) (Y) channel (c) PST (d) PCET (e) PCT Figure 5: Reconstruction of the (Y) Luminance channel of input image using PST, PCET and PCT transforms with |p| <= 20 and |q| <= 20. From the figure it can be seen that the transforms are able to reconstruct the boundary and shape of the skin lesion. 3.1 Performance of Hybrid features After training two models, with the first one using only deep features and the second on hybrid features, we compare the performance of these two models on the same testing dataset. The ISBI’s 2017 skin cancer detection challenge adopts the Area Under an ROC Curve of Sensitivity (True Positive Rate) vs. Specificity (False Positive Rate) as the performance metric. We use the same metric to compare the performance of the two approaches. Obtained ROC curve is shown in Figure 7. From Figure 7 it can be observed that the AUC increases as we increase the number of PHT transform coefficients. At the time of writing this paper, the performance of the top ten methods of the Melanoma classification in the ISBI 2017 skin cancer challenge ranges between 0.78 and 0.87 in terms of Area Under the Curve. Because we introduce the hybrid features even a lightweight network model, such as MobileNet, performs well and attains a performance comparable as some of the top ten algorithms that have predominately used ensemble of deep neural networks.20 3.2 Training time In this section, we compare the training and inference time of two models on a Raspberry Pi device. Table 2 compares the training time of the predictions models with and without the hybrid features. We also conducted experiments on an Intel Movidius Neural Computing stick (NCS) that supports the hardware acceleration for inference and measured the speedup for the model inference. Results are summarized in Table 3. We also measured the distribution of time spent in each operation to identify the bottleneck operation that needs further optimization for better performance in Figure 8. The PHT feature extraction is the slowest
7 .Figure 6: ROC curve obtained on validation set using linear, polynomial, sigmoid and radial basis function kernels on hybrid features with C = 20 for SVM. We select the radial basis function because it has the best performance. Figure 7: ROC curve obtained using only Mobile Net features and Hybrid features with varying values of C where |q| <= C and |p| <= C. operation and takes the dominant share of latency in the whole process. To ensure real-time inference, we use the Movidius stick that provides a viable option in our application. Because PHT is essentially a convolution operation with the kernel size that is equal to the size of an image, we can seamlessly deploy the PHT on an Intel Movidius stick by converting it into a neural network with the filter weights defined by the 2-D tensors of PCT, PCET, and PST. Note that the Intel Movidius has native support of Google Tensorflow. In our future work we plan to deploy the entire PHT to Intel Movidius to obtain the benefit of hardware acceleration. 4. CONCLUSION Traditional machine learning and deep neural often demands high-performance computing that is only available at the external requiring supercomputers and cloud computing. The external execution of deep learning is usually labor-intensive, involves human-in-the-loop for job submission, tracking, and monitoring, and often requires the expertise that can not be easily sustained, replaced, or scaled, especially when we approach the era of the Internet of Things (IoT). In this paper, we designed a hybrid approach that pushes the deep learning task into the edge of the Internet where network connectivity is scarce, data is confined at the local facility due to the resource constraints and data privacy concern, and a decision must be made locally with input data because of the time requirement. When a large amount of data is available, we expect the deep learning models to perform even better.
8 . Table 2: Time Comparison for training of SVM model on Raspberry Pi in minutes. Hybrid feature model (C=20) MobileNet feature model 14.02 2.79 Table 3: Time Comparison for Deep feature extraction using MobileNet for a single image in seconds. Raspberry PI Raspberry PI with Movidius NCS 2.67 0.063 Figure 8: Time taken in each operation of interference on a Raspberry Pi using hybrid features when C = 10. ACKNOWLEDGMENTS This research is partially supported by National Science Foundation Grant NSF IIS-1715985 and Brookhaven National Laboratory, NY. We also thank the Center of Excellence in Wireless and Information Technology (CEWIT) at Stony Brook University for providing us with the initial training related to using Raspberry PI. REFERENCES [1] Guy, G. P., Machlin, S. R., Ekwueme, D. U., and Yabroff, K. R., [Prevalence and Costs of Skin Cancer Treatment in the U.S., 2002-2006 and 2007-2011], vol. 48, American Journal of Preventive Medicine (2015). [2] Jain, S., jagtap, V., and Pisea, N., “Computer aided melanoma skin cancer detection using image process- ing,” in [International Conference on Intelligent Computing, Communication and Convergence (ICCC) ], 48, 735–740 (2015). [3] Bakheet, S., “An svm framework for malignant melanoma detection based on optimized hog features,” in [Computation], 5 (2017). [4] Satheesha, T. Y., Satyanarayana, D., Prasad, M. N. G., and Dhruve, K. D., [Melanoma Is Skin Deep: A 3D Reconstruction Technique for Computerized Dermoscopic Skin Lesion Classification], vol. 5, IEEE Journal of Translational Engineering in Health and Medicine (2017). [5] Yuan, X., Yang, Z., Zouridakis, G., and Mullani, N., “Svm-based texture classification and application to early melanoma detection,” in [IEEE EMBS Annual International Conference ], (2006). [6] A.Sheha, M., S.Mabrouk, M., and Sharawy, A., [Automatic Detection of Melanoma Skin Cancer using Texture Analysis ], vol. 42, International Journal of Computer Applications (2012). [7] Situ, N., Yuan, X., Chen, J., and Zouridakis, G., “Malignant melanoma detection by bag-of-features classi- fication,” in [IEEE EMBS Conference ], (2008). [8] Barata, C., Ruela, M., Francisco, M., Mendonca, T., and Marques, J. S., [Two Systems for the Detection of Melanomas in Dermoscopy Images Using Texture and Color Features ], vol. 8, IEEE Systems Journal (2014).
9 . [9] “Danger Signs: ABCDE Rule for Melanoma Detection.” Online Article https://www.txid.org/?page_id= 3412. [10] Howard, A. G. and Menglong Zhu, Bo Chen, D. K. W. W. T. W. M. A. H. A., “Mobilenets: Efficient convolutional neural networks for mobile vision applications,” (2017). [11] “Intel shows off the movidius myriad x, a computer vision chip with deep learning baked-in.” Online Article https://techcrunch.com/2017/08/28/ intel-shows-off-the-movidius-myriad-x-a-computer-version-chip-with-some-serious-brains. [12] Deng, J., Dong, W., Socher, R., Li, L.-J., Li, K., and Fei-Fei, L., “ImageNet: A Large-Scale Hierarchical Image Database,” in [CVPR09 ], (2009). [13] Sahu, P., Qin, H., and Yu, D., “Lightweight deep learning on smartphone for early detection of skin cancer,” in [Symposium on Data Science for Healthcare (DaSH), Ridgefield, Connecticut, United States, October 19- 20, 2017], (2017). [14] Esteva, A., Kuprel, B., Novoa, R. A., Ko, J., Swetter, S. M., Blau, H. M., and Thrun, S., [Dermatologist-level classification of skin cancer with deep neural networks], nature (2016). [15] Szegedy, C., Liu, W., Jia, Y., Sermanet, P., Reed, S., Anguelov, D., Erhan, D., and Vanhoucke, V., “Going deeper with convolutions,” in [CVPR ], (2015). [16] He, K., Zhang, X., Ren, S., and Sun, J., “Deep residual learning for image recognition,” in [CVPR ], (2015). [17] Yap, P.-T., Jiang, X., and Kot, A. C., [Two-Dimensional Polar Harmonic Transforms for Invariant Image Representation], vol. 32, IEEE Transactions On Pattern Analysis And Machine Intelligence (2010). [18] Moody, M. F., “Chapter 6 - two-dimensional {FTs},” in [Structural Biology Using Electrons and X-rays], Moody, M. F., ed., 95 – 121, Academic Press, Boston (2011). [19] Liu, M., Jiang, X., Kot, A. C., and Yap, P.-T., [Application of Polar Harmonic Transforms to Fingerprint Classification], World Scientific (2011). [20] Codella, N. C. F., Gutman, D., and et al., M. E. C., “Skin lesion analysis toward melanoma detection: A challenge at the 2017 international symposium on biomedical imaging (isbi), hosted by the international skin imaging collaboration (isic),” in [ArXiv ], (2017).